
Animal Complexity
What determines animal complexity? What about humans makes them more complex than plants? Differences in species complexity was originally thought to be caused by different amounts of DNA, where yeast, a simple organism, would have less DNA, and more complex beings, like humans, would have more. By having more DNA, it was also thought that more complex organisms would then have more proteins. This is both true and false, leading to a phenomenon called the C-value paradox. The C-value paradox describes the lack of correlation between genome size and protein coding genes. Take a look at the chart below. The green bars in the top represent the size of the genome (amount of DNA) in various organisms, from yeast to humans. If we now look at the bottom chart in purple, which represents the estimated amount of protein genes in each organism, yeast still has the fewest. However, the organism with the most protein coding genes is the parasite Trichomoniasis. Even rice has more protein coding genes than humans!
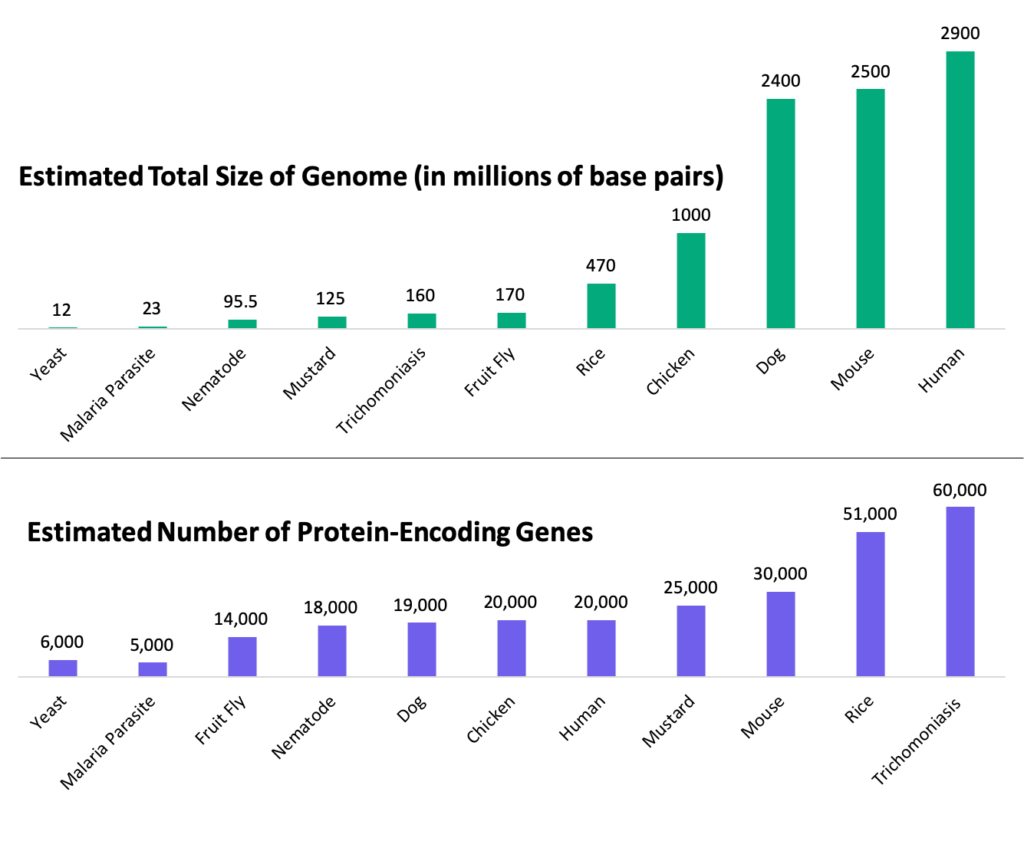
Bottom: estimated number of protein coding genes of various organisms
Estimates as of 2008.
So while genome size does correlate with organismal complexity, the number of protein coding genes doesn’t. Rather, what correlates with organismal complexity is the size of an organism’s non-coding genome, or the part of the DNA that doesn’t have the ability to become protein.
DNA is transcribed into RNA, and then RNA is translated into protein.
The Central Dogma of Biology
The central dogma of biology (figure to right), first proposed by Francis Crick in the 1960s, states that DNA is a template that is transcribed into an RNA intermediate, which is finally translated into protein, the functional unit in our cells. During the decades since the proposal of the central dogma, we have learned that most of our genome doesn’t become protein, and such parts of the genome were thought of as “junk DNA,” useless sequence left over from evolution. However, we now know that proteins aren’t the only molecules in our bodies that have functions. RNA can also have functions other than becoming protein. These RNAs that do not get translated into protein are called non-coding RNAs.
What do Non-Coding RNAs do?
Non-coding RNAs do a lot of things! Their main function is regulating gene expression, but they have many other functions as well, like helping with translation and organizing DNA in the nucleus. Because of these different functions, there are many classes of non-coding RNAs. Transfer RNAs, or tRNAs, help build proteins by carrying the amino acids that will be linked together. Ribosomal RNAs, or rRNAs, interact with ribosomes to carry out the process of translation. In terms of gene expression regulation, an endless list of classes of non-coding RNAs could be described, but some of the larger classes include micro RNAs (miRNAs), which interact with mRNA prevent protein production through a process known as RNA interference (RNAi), and long non-coding RNAs (lncRNAs), which interact with proteins to turn genes on or off. How does organismal complexity relate to non-coding RNAs? Non-coding RNAs tend to have the highest expression in brain tissue, suggesting that gene expression in the brain is very precisely regulated – allowing for complex and abstract thought, characteristics we attribute to complex organisms, such as human beings.
Non-Coding RNAs in Action: X Chromosome Inactivation

Sex chromosomes determine whether organisms will develop into males or females. Normally in mammals, males have an X and a Y sex chromosome, and females have two Xs. This means that females have twice as much X chromosome DNA as males. To make up for this difference, one of the female Xs is randomly turned off early in development in a process called X Chromosome Inactivation (XCI). XCI occurs when a lncRNA brings the protein PRC2 to the X chromosome. Once on the X chromosome, PRC2 turns off genes, through the addition of epigenetic marks. The effects of XCI are best exemplified in calico/tortoiseshell cats, since the gene that controls fur color is on the X chromosome. If the X chromosome with the gene that makes orange fur is turned off, then that cell makes black fur, and all of the daughter cells will also make black fur. Depending on which X gets turned off and at what time in development determines which fur colors and patterns the cat will have.
Non-Coding RNA Research at UNC
Understanding the complexity of our genome (all DNA) and transcriptome (all RNAs) is an increasingly active area of research and will become pertinent for the development of future therapeutics. The function of non-coding RNAs comes from the proteins with which non-coding RNAs interact. Studying both different non-coding RNAs, as well as the proteins these non-coding RNAs interact with, are crucial pieces to the puzzle of biology.
The University of North Carolina at Chapel Hill is lucky to have diverse expertise in non-coding RNA and RNA Binding Proteins:
– It is estimated that humans have up to 20,000 lncRNA genes, but only one of these lncRNAs, the one involved in XCI, has been extensively studied. The lab of Dr. Mauro Calabrese studies how lncRNAs like the one involved in XCI turn off genes.
– Humans have hundreds proteins that are able to interact with and regulate RNA. Dr. Daniel Dominguez studies the properties of these RNA binding proteins and how these proteins cause human diseases.
– Animal complexity can also be partially explained through alternative splicing, which is the concept that one gene can be made into different proteins. The lab of Dr. Jimena Giudice focuses on alternative splicing and the RNA binding proteins involved in these processes.
– Non-coding RNAs can also help increase gene expression – these RNAs are called enhancer RNAs. Dr. Hector Franco is interested in understanding the roles of enhancer RNAs in cancer.
– RNA can fold into 3-D structures, and these structures determine what proteins can bind to the RNA. Dr. Alain Laederach uses computational and experimental techniques to understand the roles of RNA structure in cell processes.
… and many, many more.
Peer edited by Matthew Tsilimigras
One Reply to “What Determines Our Complexity?”