
Scientists thrive on “aha” moments— breakthroughs in knowledge that come from careful planning or perhaps fortuitous luck. For a team of researchers led by Josh Lawrimore, a fourth-year graduate student in Kerry Bloom’s lab at UNC, their “aha” moment came about by approaching their research question in a new way.
Josh’s research is focused on what happens to a chromosome—a long molecule of DNA wrapped around proteins—when a parent cell divides into two daughter cells. To study chromosomes, the Bloom lab uses baker’s yeast as an experimental model. The centromere region in yeast cells has been well-studied, and their chromosomes are similar in structure to human chromosomes. Amazingly, through working with this simple organism, Josh has solved a long-standing mystery in the field of cell biology.
No Chromosome Left Behind
Josh explains that cell division, an essential process for growth and development, must happen without mistakes, “otherwise, you get chromosome instability, which is, of course, a hallmark of cancer”.
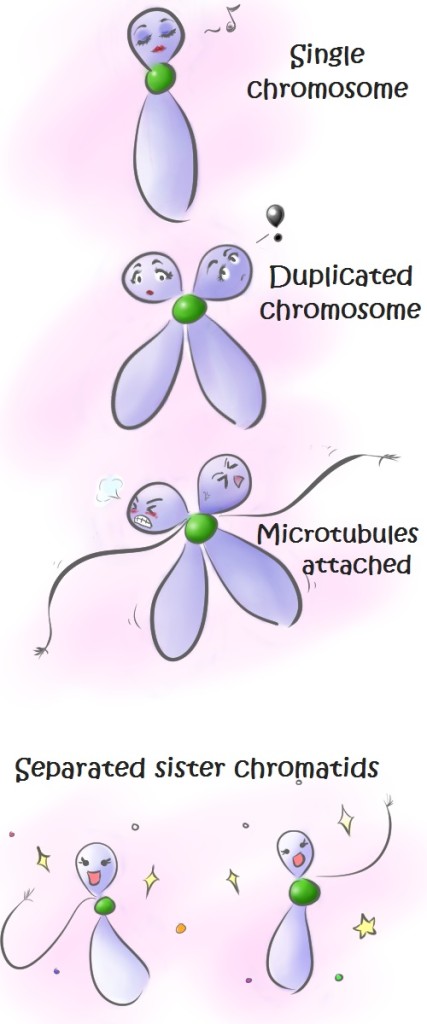
In a classical model of cell division (see image and caption above), proteins called cohesins hold sister chromatids together until they separate at later stages of cell division. When microtubules attach to the centromere, they generate tension against the cohesin located there. This tension sends a signal to the cell that microtubules are correctly attached to all chromosomes. Then, sister chromatids are separated.
“But, if you look under the microscope” Josh explains, “there is no persistent sister-sister cohesion at the centromere.” So, what generates tension prior to separation? And, why is cohesin, which holds the chromatids together, located at the region of separated centromeres?
The Bloom lab, in addition to the Mitsuhiro Yanagida and Peter Sorger labs, published this paradoxical finding about ten years ago. However, it didn’t immediately change the prevailing model of cell division because each group had its own interpretation. For his dissertation work, Josh set out to gain insight into this baffling observation.
Another observation, published several years later, provided an important clue. Chromatin adjacent to the centromere forms multiple outward-facing loops sometimes called a bottle brush, similar to the first step of tying shoelaces. DNA looping has long been known to bring distant sites on DNA into close proximity for different functions. The Bloom lab wondered if these chromatin loops might contribute to proper sister chromatid separation.
Cell Biology Meets Polymer Physics
Josh explains that, less than ten years ago, the field of cell biology realized chromosomes behave like simple polymers. He reasoned that approaching his questions from a biophysical standpoint would provide more detail than simply observing the chromosomes under a microscope. Josh consulted with scientists Greg Forest and Paula Vasquez at the Carolina Center For Interdisciplinary Applied Mathematics. They explained that, based on polymer physics principles, the observed bottle brush organization of chromatin would create tension in the same direction as microtubule pulling.
Josh tested this prediction experimentally, and his results revealed, “the loops themselves can generate an outward force in addition to the [microtubule] pulling forces.” When he inhibited microtubule function, Josh further explains, he found “microtubules only [produce] about twenty percent of the motion you see instead of one-hundred percent.” That the movement of chromatids during chromosome separation was not solely from pulling by microtubules was a big surprise. This result revealed how tension could be generated at the centromeres, given the distance between them.
Cell Biology Meets Computer Science
From these studies, published in 2015, the Bloom lab also learned that the presence of cohesin (and another protein, condensin) help organize the elaborate loop configuration. This observation revealed why cohesin was located at the centromere region. However, their results begged the question, why does the chromatin form all those loops when the simple model of holding the sisters together with cohesin makes more sense?
From a polymer physics standpoint, Josh learned that the loops generate tension by virtue of their thermodynamic properties: they bump into and repel each other. Because these properties dictate structural dynamics, and because determining structure aids in understanding function, Josh realized that defining the thermodynamic underpinnings of their system could help fill in the details of this new view of cell division.
The Bloom lab (including UNC undergraduate students Patrick Hahn, Ben Kompa, and Alyona Fulp) and their collaborators at the UNC Center for Computer Integrated Systems for Microscopy and Manipulation (Russ Taylor II, Michael Falvo, and Leandra Vicci) made a three-dimensional model of the centromere. The model was based on their experimental observations of the size and distribution of cohesin and condensin and the structural properties of DNA. They could then simulate, or predict, how the three-dimensional centromere organization could affect its structure in the real-life cellular environment. They named the mathematical simulation program ChromoShake (published earlier this year).
The computer simulation experiments predicted that the outward force exerted by the rings causes the chromatin at the centromere region to become stiff, which likely aids the microtubules in getting a firm grip on the DNA. Importantly, when the Bloom lab performed experiments to validate the computational predictions, Josh says “the simulated data and experimental data are remarkably similar.” This emphasizes the ability of their models to accurately predict the function of chromosome structural components.
The Big Picture
Other aspects of Josh’s dissertation work provide additional information on chromosome structural dynamics. A loop or loops can collapse to release tension when microtubules pull hard enough, and then the loops reform once the microtubules give slack. Thus, a dynamic balance of tension occurs at the centromere, not a hold them together tight then pull them apart mechanism.
In contemplating these results, Josh recalls the sage words of his graduate advisor, Kerry Bloom:

“If you look at the efficiency of chromosome segregation, it has a failure rate of somewhere between one in one-hundred thousand to one in a million. No human machine that I know of…operates at that level of efficiency. The principles of thermodynamics explain how that kind of efficiency is obtained.”
The marriage between biology and physics happened over a hundred years ago, demonstrating how physics principles underpin the structural and functional properties of the cellular environment. Josh’s dissertation work further demonstrates how taking into account invisible forces described by physics principles provides information not available using only his favorite tool, the microscope. When asked if these principles are applicable to other aspects of cell biology, Josh replied, “that’s where we’re headed next.”
For more information on Josh’s work, see his recent publications describing the chromatin loops and the Chromoshake simulator. For more information on the Bloom lab’s other research, visit their lab website.
Peer edited by Michael Clark and Amy Rydeen
Illustration by Lindsay Walton
Follow us on social media and never miss an article featuring a UNC Scientist:
One Reply to “Biology and Physics Meet in the Middle”