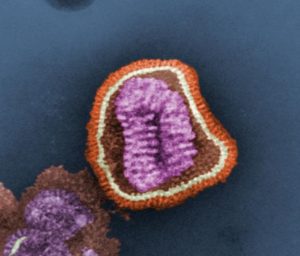
Influenza is a virus that straddles two worlds: that of the past and that of the future. Responsible for more deaths than HIV/AIDS in the past century, the flu is one of the world’s’ most dangerous infectious diseases though it may not seem so, especially in the United States. However, the flu is responsible for millions of cases of severe illness and approximately 250,000 to 500,000 deaths worldwide each year.
Influenza Pandemics
Influenza A and B circulate each flu season, but it is the emergence of new influenza A strains that have been responsible for worldwide epidemics, or pandemics, in the past such as the 1918 ‘Spanish Flu’ pandemic and the 2009 H1N1 pandemic. There are 2 ways a new influenza virus can emerge. Every time the virus replicates, small genetic changes occur that result in non-identical but similar flu viruses: this is called “antigenic drift”. If you get infected with a certain flu virus, or get a vaccine targeting a certain flu virus, your body develops antibodies to that virus. With accumulating changes, these antibodies won’t work against the new changed virus, and the person can be infected again. The other source of change is “antigenic shift”, which results in a virus with a different type of hemagglutinin and/or neuraminidase, such as H3N2 to H1N1. The 2009 H1N1 virus is cited by some as a result of antigenic shift, because the virus was so different than previous H1N1 subtypes; however, as there was no change in the actual hemagglutinin or neuraminidase proteins, it was technically a case of antigenic drift.
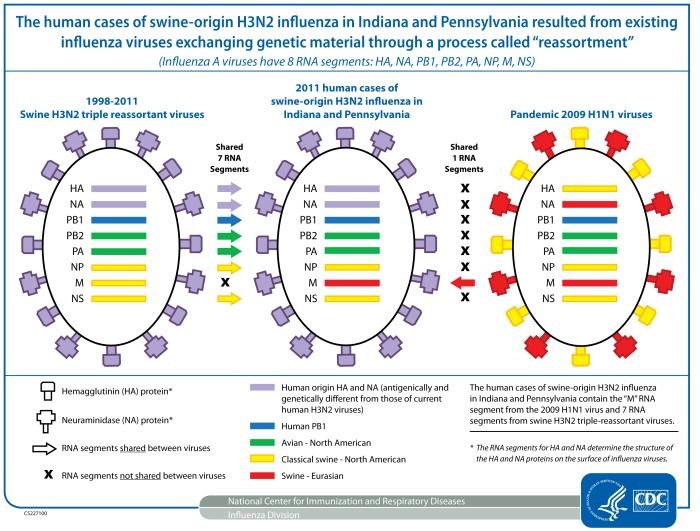
Content Provider: CDC/Douglas Jordan, M.A. 2011.
Challenges in Studying the Flu
Scientists and policymakers face many challenges when studying the influenza virus. For instance, the virus can be transmitted among people not showing symptoms and cough, sneeze, or handshake can spread infectious droplets from someone who doesn’t know they’re sick. Scientific study is further complicated by the virus itself: there are 3 antigenic types of influenza virus that infect humans, (A, B, C), with various subtypes and strains. Each year, government agencies work with scientists to decide which strains to target in that year’s vaccine manufacturing. The lag time between production in the spring and the flu season in the winter provides time for unexpected types to emerge.
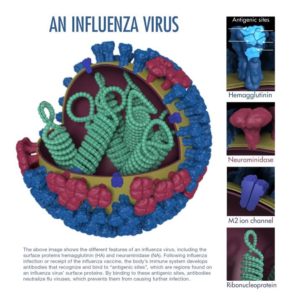
The H#N# nomenclature for influenza A subtypes refers to the hemagglutinin (H) and neuraminidase (N) proteins that sit on the surface of the virus. There are 18 types of hemagglutinin and 11 types of neuraminidase. Hemagglutinin aids the virus in fusing with host cells and emptying the virus’ contents inside. Neuraminidase is an enzyme embedded in the virus membrane that facilitates newly synthesized viruses to be released from the host cells to spread the infection from one cell to another.
Targeted Therapy
In studying ways to prevent and battle influenza, research scientists have focused their efforts on blocking the actions of neuraminidase and hemagglutinin. Antiviral drugs, such as oseltamivir (Tamiflu®) and zanamivir (Relenza®), bind neuraminidase, both interact with neuraminidase at sites crucial for its activity. The drugs act to render the virus incapable of self-propagating. A computational biologist at the University of Washington in Seattle, David Baker and his team know the hemagglutinin protein well. In 2011, they utilized nature’s design by studying antibodies that bind hemagglutinin in order to design a protein that targets the glycoprotein’s stem in H1 subtype flu viruses and prevent the virion from infecting the host cell. However, antiviral resistance contributed to by antigenic drift, is a serious issue. Researchers much constantly develop new drugs to keep up with changes in the virus.
David Baker and his team now focus their research on the hemagglutinin protein. Utilizing a computational biology approach, they designed a protein that fits snugly into hemagglutinin’s binding sites. They tested their designer protein on 10 mice and found that in mice exposed to the H3N2 influenza virus, their protein worked both as a preventative measure and as a treatment. Though there is a long road to human testing, this binding protein shows promise for bedside influenza diagnosis as well as a model for possible treatments.
Want to know more?
Learn how scientists monitor circulating influenza types and create new vaccines each year.
See flu activity and surveillance efforts with the CDC’s FluView and vaccination trends for the United States using the FluVaxView.
Peer edited by Richard Hodge and Tyler Farnsworth.
Follow us on social media and never miss an article: