When you think of the word “protein” you probably think of things like steaks and protein shakes. However, when biologists talk about proteins they mean the molecules that are abundant in our bodies and carry out numerous functions. These protein molecules are made of smaller building blocks called amino acids. The sequence of amino acids in a protein defines its shape, which in turn affects its function.
Scientists can both manipulate the structure of naturally-occurring proteins and design entirely new protein structures through computational protein design. Computational protein design involves designing a sequence of amino acids that is predicted to fold into the desired structure.
The traditional process of designing proteins from scratch requires knowledge of the rules behind how certain protein structures fold, which isn’t as easy as it seems. Dubbed “the protein folding problem”, scientists have been trying to figure out how proteins fold for decades. In nature, a protein is usually found in its lowest energy shape. Even though amino acids can rotate in many ways – resulting in countless possible structures – as long as the amino acid sequence and environmental conditions stay the same, the protein will fold the same way nearly every time. The desire to solve the protein folding problem has guided the development of computational tools to predict protein structure. Thanks to success from DeepMind’s machine learning algorithm called AlphaFold, scientists are closer than ever to obtaining near-experimental accuracy for protein structure prediction using machine learning.
Moreover, machine learning techniques are also used for protein design. For instance, researchers at the University of Washington recently published a paper describing a new technique called “deep network hallucination” that utilizes a type of machine learning called deep neural networks to generate completely new proteins from random amino acid sequences. Deep network hallucination eliminates the need for prior knowledge of the desired protein fold which makes the design process easier.
The neural network used for “hallucinating” protein designs is called trRosetta and is trained using experimentally-determined protein structures found in nature. Using these native structures, the model is able to learn what amino acid positions and orientations define “good” protein shape. trRosetta can be used to predict structure when given the sequence of a naturally occurring protein. By contrast, the hallucination method involves giving the neural network a random sequence of amino acids. Eventually, the network outputs protein structures that are called “hallucinations” because they are dreamt up by the network. This methodology is similar to the types of algorithms that can generate entirely new human faces.
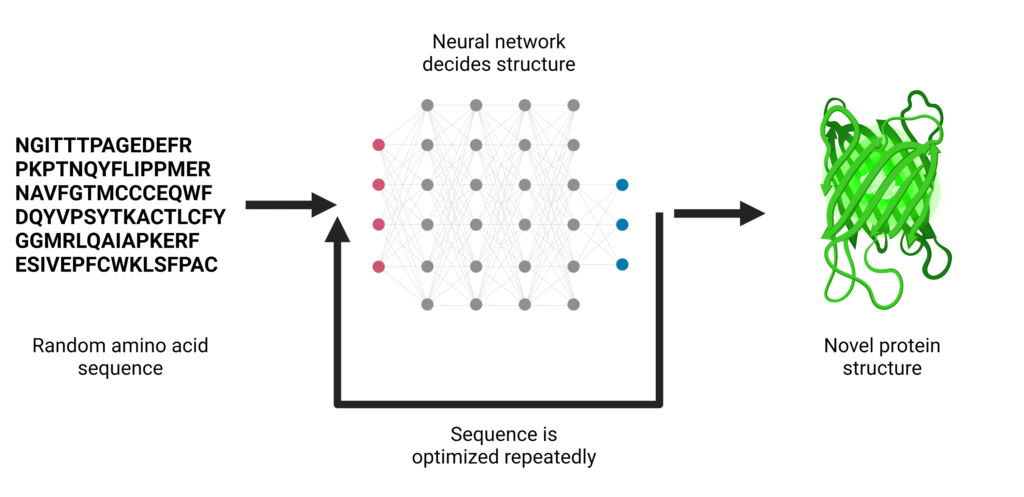
After generating thousands of hallucinated protein structures, the University of Washington research team experimentally tested over 100 of the hallucinated designs in the lab. Several of their designs aligned closely with the computational hallucinations. The close agreement between the experimentally-solved structures and the hallucinated designs indicates that the hallucination method is capable of producing entirely new proteins, including some designs with protein shapes that have never been found in nature.
Using machine learning to design proteins is still a new concept. But as the fields of both machine learning and protein design continue to advance, it will surely prove to be a powerful tool that has the potential to revolutionize the field of structural biology.
In fact, work has already started that utilizes machine learning methods such as AlphaFold and the similar RoseTTAFold to design new proteins. A recent preprint article described a novel method that uses AlphaFold to predict the structure of random amino acid sequences. The authors showed success in designing proteins called binders that adhere to specific target proteins. Such binders are commonly used in the development of diagnostic and therapeutic tools. Therefore, a machine learning design framework could enable rapid and efficient drug and therapeutic design.
Header image shows 3 of the hallucinated protein structures. 7M5T in green (left), 7M0Q in pink (middle), and 7K3H in blue (right). Figure created using PyMOL by Mikayla Feldbauer.
Peer editor: Jeanne-Marie McPherson