New research shows proof of concept for a potential class of drugs that could inhibit disease-causing proteins by tearing them to shreds.
The findings, published in Nature Chemical Biology in December 2022, describe the discovery and validation of a peptidic chemical compound that can direct proteins to their own degradation. In essence, it brings misbehaving proteins straight to the mouth of an intracellular protein shredder—called the proteasome—which naturally exists in our cells to maintain balance or homeostasis.
Drugs that degrade a protein could make a single dose last significantly longer than those that simply inhibit protein function, which is how most drugs work nowadays. If a twice-daily pill became a once-weekly pill, drug manufacturing costs (and presumably market price) could decrease. At the same time, patient compliance might increase, and further still, historically challenging protein targets with unclear functions could become ‘druggable’ through their degradation.
Researchers from Genentech Inc. in South San Francisco used a peptide drug discovery screening tool called mRNA display to discover this director of protein destruction. This tool searches through up to ten trillion peptide sequences to identify those that bind to a selected target. Because the researchers wanted to bait aberrant proteins to the proteasome, they chose to perform the mRNA display screen against a part of the proteasome called PSMD2.
The researchers identified a handful of peptides that bound tightly to PSMD2 alone and further confirmed that most could also bind PSMD2 in the whole proteasome. They then used cryogenic electron microscopy to observe the structure of their top peptide, MC1, bound to PSMD2, and observed that MC1 was located relatively close to the mouth of the proteasome, where proteins are fed for degradation.
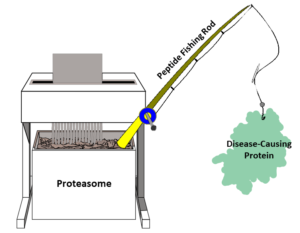
Linking the proteasome binder MC1 to another bait ligand of a misbehaving protein could then lure the protein to the proteasome for degradation. In this sense, MC1 acts much like a fishing rod to which scientists can attach various types of bait to ‘catch’ different proteins.
As proof of concept, the researchers chose to bait a protein called BRD4, which regulates gene expression and is overactive in various cancers. They linked the bait ligand of BRD4 to MC1 and, after optimizing the placement and length of the linker, showed that it could bring BRD4 to the proteasome to cause its degradation in human kidney cells.
BRD4 was chosen as a model protein as other researchers have previously baited it in an alternate protein degradation approach using proteolysis targeting chimeras, or PROTACs for short. A PROTAC would recruit BRD4 to specific enzymes that mark it with multiple tags that the proteasome can recognize. PROTACs have garnered significant research interest, and several have entered clinical trials.
However, as this recent report highlights, PROTACs aren’t without limitations. Unlike the proteasome, which is present in all cells, the enzymes that PROTACs rely on to ‘tag’ misbehaving proteins for degradation are not. Additionally, any proteins selected for PROTAC-induced degradation must have specific handles on their surface for these tags to attach to. These requirements restrict PROTAC utility to certain tissues and proteins in the body. In contrast, the present study circumvents the need for ‘tagging’ altogether by simply bringing the protein directly to the proteasome for degradation.
Though much future work remains, MC1 shows promise as a multifaceted ‘fishing rod’ that can lure misbehaving proteins directly to the proteasome to meet their demise. With any luck, these direct degraders may circumvent the limitations of PROTACs and create a more versatile, ubiquitous, and powerful therapeutic class.
Peer Editor: Bhavyaa Tyagi