Not many people think about the complex regulation of digestion, or in other words how we gain nutrients from the food we eat and rid ourselves of unnecessary (and sometimes toxic) byproducts.
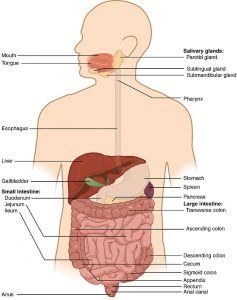
More specifically, we first put food in our mouth and use a combination of mechanical (chewing) and chemical (the enzymes, or proteins that act like catalysts, in saliva) forces to breakdown our food into smaller pieces, where it gets pushed through the esophagus through muscle contractions to the stomach. The stomach contains enzymes that break down food even further, where it is then passed into the small intestine.
The small intestine contains three separate subsections: the duodenum, which receives the food from the stomach, bile from the gallbladder, and enzymes from the pancreas to finish digestion; the jejunum, where most of the digested food is absorbed into the bloodstream; and the ileum, where some of the remaining nutrients are absorbed and the byproducts get passed to the large intestine. The large intestine consists of five separate subsections, including the cecum, (which absorbs water and salts), and the four main sections of the large intestine (the ascending, descending, transverse, and sigmoid colon), where it absorbs any remaining water and nutrients, causing it to solidify into feces. The descending colon stores the feces, and the sigmoid colon contracts to increase the pressure inside of it, causing the feces to go into the rectum. Finally, the rectum holds the feces awaiting elimination by defecation. The sections of the small intestine and large intestine are collectively known as the gut.
In the last couple of years, numerous studies have explored the function of our gut system, with the greatest focus on how the gut’s legions of bacteria, viruses, and other microbes (also known as the microbiome) interact with our gut. But the discoveries in these sectors often overshadow the fact that we are still learning about what happens in the gut (and other tissues, really) at the cellular level. That said, over the past few years there has been a large multi-lab, multi-institutional initiative called the Human Cell Atlas that seeks to map and chart the course of every cell type in the human body from birth to death, as a reference point from which to study when and how cell type activities are altered during different periods of development and in different diseases.
In a paper published earlier this month, Elmentaite et. al. made a major contribution to this effort by isolating cells from multiple tissues of the second-trimester fetus and adult guts, taking the RNA from these cells, barcoding the RNA molecules so we know which cells they came from, then sequencing the genetic information; this process is known as single-cell RNA sequencing (or scRNA-seq). They also added data from a prior study where they took intestinal tissue from first-trimester fetuses, the small intestinal tissue from children with a chronic intestinal inflammatory condition called Crohn’s disease, and the small intestinal tissue from healthy children (see this paper). This led to them collecting a total of more than 428,000 high-quality cells, and through a process known as clustering (where they place cells into groups based on similarities in gene expression patterns), found 10 major types of cells (including cells making up different layers of the intestine, blood cells, and immune cells). They showed that the composition of those cell types were different both through developmental time and in Crohn’s Disease, and that the gene expression patterns in the small and large intestine overall were different between children and adults. Further clustering among each of those major cell types led to the discovery of 133 different cell types. From there, they further characterized the cells that made up the intestinal epithelium, the enteric nervous system, and the lymphatic system, by mapping both their developmental trajectories and gene expression changes through development. They were able to show novel findings in each of these three systems (which will be expanded upon more in the subsequent paragraphs).

The intestinal epithelium is the outer lining of the intestine, which is between the immune cells in the intestine (in the lamina propria) and the microbiota outside of it (the lumen). If the cells are being attacked by foreign microbes, viruses, or other pathogens, the immune cells can launch an immune response that will fight back against the invader. The layer of cells that make up the epithelium is known as the intestinal crypt. At the bottom of the crypt are the stem cells (which can divide to make more of themselves and can differentiate into more mature cells) and Paneth cells (which contain antimicrobial peptides that can protect against microbial attack). As you go further up the crypt you encounter enteroendocrine cells (which produces peptide hormones that regulate metabolism), goblet cells (which produces a sticky layer of mucus on the surface of the cell that forms a barrier between the immune cells and the microbiota), enterocytes/colonocytes (which take antigens, or molecules found on the surface of the pathogens, from the lumen and present them to immune cells in the lamina propria, thus activating an immune response), a normally rare cell type known as the tuft cells, and a newly discovered enterocyte/colonocyte subtype known as BEST4 cells.
Through this scRNA-seq analysis, they learned that a gene known as PLCG2 was expressed by these tuft cells and at higher levels than in certain immune cells. This gene was associated with another gene, FCGR2A, that is known to be turned on in response to immune stimulation, suggesting that tuft cells might play a role in responding to immune cell signaling. Furthermore, different gene expression patterns in BEST4 cells in the small intestine in comparison to the large intestine suggested that BEST4 cells in the small intestine play a role in helping goblet cells produce mucus and producing acid, while BEST4 cells in the large intestine play a role in the metabolism of small molecules.
Furthermore, they looked at the cellular makeup of the enteric nervous system. While many people know that the brain is composed of cells known as neurons (which receive and transmit electrical signals to target cells) and glial cells (which broadly speaking provide support to the neurons), it is less well known that the intestine has these cells as well, and that these cells both receive instructions from the brain and pass along information to the brain, through a connection system known as the gut-brain axis. Through scRNA-seq, they mapped the different differentiation pathways of neurons and glia at different stages of fetal development. They followed the differentiation path of a progenitor of neurons and glia known as enteric neural crest cell progenitors (ENCC), and showed at the earlier time point that the ENCCs predominantly became different types of neurons, while at later time points they differentiated into both glial and neuronal cell types. They also looked at the gene expression patterns of genes known to play a role in Hirschsprung’s disease (a disease that causes issues with expelling feces), and found that these gene signatures were expressed across multiple cell types with varying degrees of intensity, showing that there is no specific cell type that can be implicated.
Lastly, the researchers looked at cells from the gut-associated lymphoid tissues and mesenteric lymph nodes, which play a role in gut immune cell’s surveillance for pathogens. The formation of these lymphoid tissues occur through interactions between mesenchymal and endothelial lymphoid tissue organizers (mLTo and eLTo, respectively) and lymphoid tissue inducers (LTi). They found gene signatures of these specific subsets at early developmental timepoints, suggesting these cellular interactions start to occur early in life.
All in all, their work is a strong addition to our body of knowledge about the cellular types that make up the gut. In some cases they confirm what we already know about the development of the gut, but many of the findings brought new discoveries to light and helped us learn more about cell types we are just beginning to discover. By understanding what happens in normal development and where cellular machinery goes wrong in diseases, we will be able to better pinpoint therapeutics that will help us detect and treat diseases.
Peer Editors: Sarah Rothstein & Caroline Yu
One Reply to “Gut Check: Understanding the Human Gut Cell Landscape and Functionality Across Development”