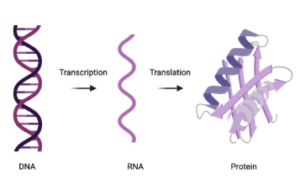
Within every cell, proteins are busy at work. They are synthesizing energy, carrying cargo from one cell to another, altering gene expression, and conducting so many other functions. Proteins are large molecules that are made up of amino acids and are the building blocks for all organisms. The central dogma of molecular biology is a fundamental biological concept that describes the flow of genetic information where DNA (deoxyribonucleic acid) transcribes into RNA (ribonucleic acid), which then translates into proteins (Figure 1). Since its establishment in the 1950s by scientists Watson and Crick, the central dogma’s function has been discovered to be far more complicated than the unidirectional schematic shown in Figure 1. With the discovery of the proteins responsible for making the central dogma flow as it does, came a new understanding of how the central dogma is altered in human disease. Whether a disease arises from a dysfunctional protein or a single nucleotide polymorphism (SNP) mutation (a mutation in DNA where one of the four DNA building blocks switches to another) – the central dogma is behind it all.
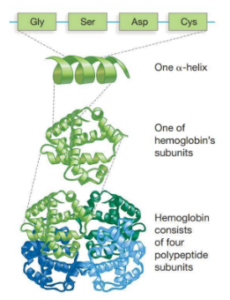
As mentioned, proteins are made up of a primary amino acid sequence, which forms localized folding patterns glued together through hydrogen bonding. The tertiary structure of proteins describes the three-dimensional shape and the quaternary structure is composed of multiple protein subunits that come together to form the fully functional protein complex (Figure 2).
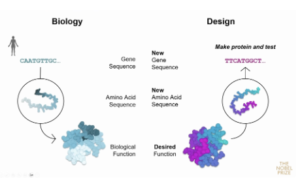
In recent years, scientists have developed strategies to create proteins that do not exist in nature from scratch, or de novo, to address modern medicine challenges that arise from disregulation of the central dogma.
In 2024, Dr. David Baker received the Nobel Prize in Chemistry, alongside Drs Demis Hassabis and John Jumper, for their contributions to protein research. Baker is currently a professor of biochemistry at the University of Washington, Seattle, where he directs the Institute for Protein Design (IPD), a research conglomerate of hundreds of trainees, multiple collaborating research groups, and clinicians.
However, before establishing the IPD and winning the Nobel Prize, Dr. Baker was a junior faculty interested in designing proteins that don’t typically exist in nature. Proteins have evolved to perform highly diverse and specialized functions but as humans live longer and acquire different and new diseases, there is a need for rapid solutions to the growing list of biological problems. Initially, Dr. Baker and his colleagues at the time, sought to accomplish their goal of de novo protein design by identifying the function of a protein they wanted to make, essentially reversing the order of events in the central dogma.
Baker and his lab at the time introduced ab initio, Latin for “from the beginning”, protein structure predictions by first computationally designing an amino acid sequence inspired by those existing in nature or modifying an existing sequence to achieve a desired structure and function. These changes may include changing a protein’s stability, reactivity, or its interaction dynamics. (Figure 3).
In 2003, David Baker and Brian Kuhlman, currently faculty in UNC’s department of biochemistry and biophysics, designed a predicted protein structure which they named “Top7” using their software called RosettaFold. They then successfully reconstructed the three-dimensional structure of Top7 using x-ray crystallography. Since this breakthrough, the Baker lab has dedicated itself to further optimizing their protein design protocol by releasing new and accessible software (RoseTTAFold, ProteinMPNN, etc.) and ultimately designing new proteins for a wide variety of applications, from vaccines to biological sensors.
AlphaFold2, invented by Drs Demis Hassabis and John Jumper (who shared the other half of the Nobel Prize with Dr. Baker) is an artificial intelligence (AI) system for protein structure prediction. With these tools, we can predict the structure of newly discovered proteins but also design new proteins to function in ways existing proteins cannot.
A recent example of designer proteins transitioning from bench to bedside is the work out of Dr. Neil King’s lab at the IPD. Long before the COVID-19 pandemic, Dr. King’s lab was developing computationally designed, self-assembling nanoparticles for a potential influenza vaccine that would potentially target multiple flu viruses. Their efforts led to a vaccine, now authorized in the United Kingdom and South Korea called SKYCovione.
More examples of real-world protein design applications are actively emerging, with designer proteins targeting diverse areas of research from climate change to drug delivery. There are some potential drawbacks however, namely the wide usage of AI systems to perform structure predictions that may not be reliable as its knowledge is limited to what it has been trained on. There are also detrimental environmental impacts of using AI, which you can learn more about here.
Despite this, it goes without saying that the future of protein structure prediction and de novo protein design offer practical, tangible, and exciting ways to revolutionize biomedicine.
Edited by: Tiffany Peters