
The pandemic has certainly gotten everyone thinking more critically about what exactly is floating in the air we breathe. From masking to outdoor gatherings to opening windows on public transit, several mitigation tactics, although divisive at times, have made it clear to the public that the spread of COVID-19 primarily occurs through the air. When airborne particles carrying respiratory viruses are inhaled, infection can occur, which is why layered, well-fitting masks are very effective at preventing the spread of COVID-19.
As technology evolves and improves our understanding of the microscopic world around us, it is becoming more clear that germs, especially respiratory viruses, are transported through particles in the air. Pollen, bacteria, dust, and smoke are other examples of particles in the air which are routinely inhaled. But what else is in the air we are breathing–and what can we learn from it?
Well–the DNA of birds, mammals, and fish–for one thing.

A recent study at the Copenhagen Zoo in Denmark in England found that eDNA from animals can be measured out of thin air. DNA, which constitutes the building blocks for all living things, can be extracted from tissues and then used to identify plants, bacteria, and animals. eDNA specifically, short for environmental DNA, is when genetic material of an organism is extracted from non-living environmental samples like water, soils, and dusts. The DNA-bearing matter that was once attached to the living thing contains enough genetic material for researchers to gather information on what, and more specifically who, left the material behind. So how is the DNA within these substances sourced back to individual organisms like plants, birds, and mammals? As it turns out, tracing can be conducted from cells and tissues that are consistently left behind by animals wherever they go, ranging from skin and saliva to feces.
Now, for the first time, eDNA has been pulled from particles in the air.
Specifically, researchers found airborne DNA which could be sourced back to 49 species of animals, including 30 mammals, 13 birds, 4 fish, 1 amphibian, and 1 reptile species. Some of the more interesting species included antelope, elephants, kangaroos, ostriches, parrots, salmon, sloths, and squirrels. The researchers hypothesized that they would detect DNA from only the zoo animals, but they were surprised to find DNA from animals not living at the zoo. DNA was also detected from local species in surrounding habitats (the squirrel) in addition to DNA from freeze-dried fish fed to several of the zoo animals (the salmon).
Interestingly, not all animals’ DNA was observed in equal amounts. The DNA from mammals with larger biomass, or a larger weight-to-size ratio, was more likely to be detected in the air when compared to smaller animals, like birds. Further, animals located closer to the sensors were also more likely to be detected, suggesting that airborne DNA can only travel a certain distance. The wind direction also played some role in the detected DNA, as only animals upwind of the sensors were detected. More research must be conducted to determine how other weather conditions, such as wind speed, sunshine, or rain, might impact the detection of airborne eDNA.
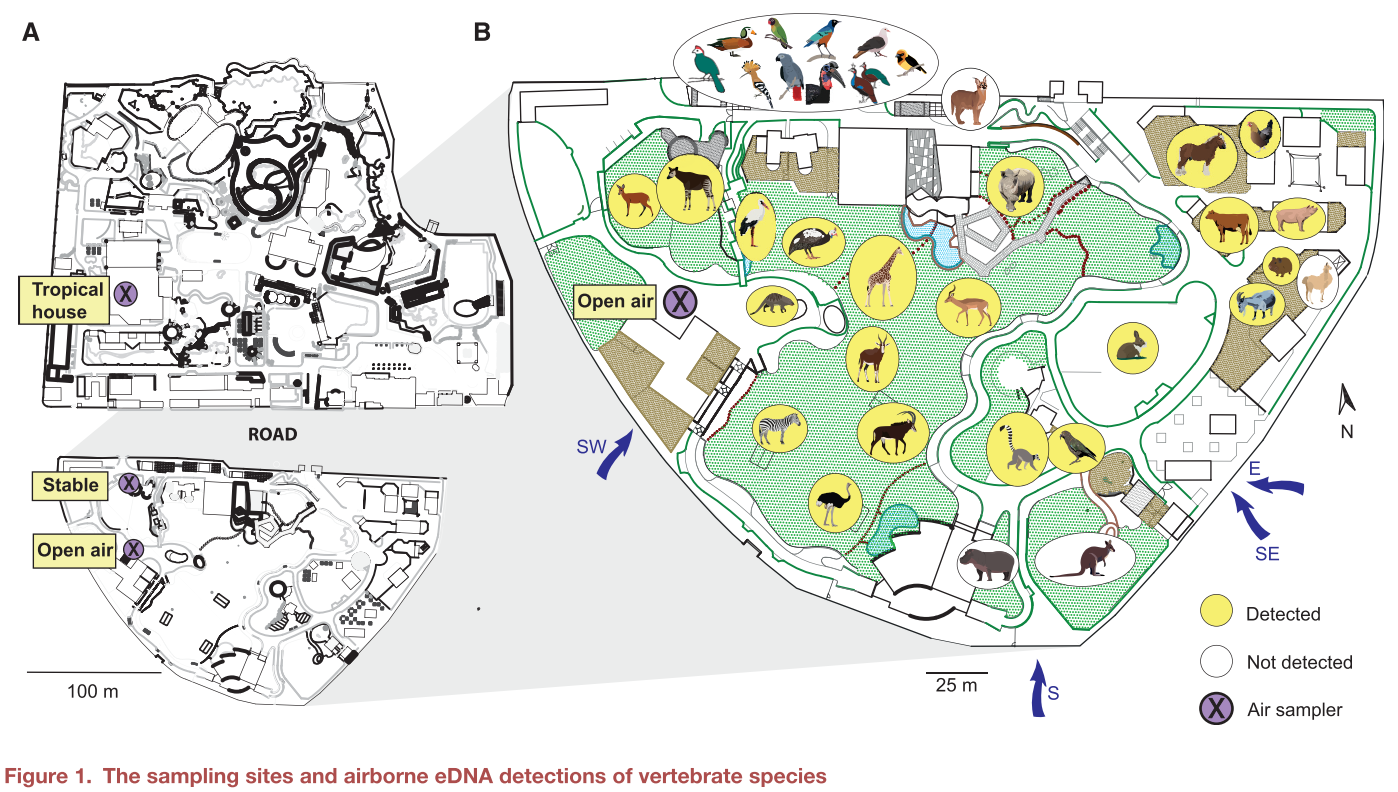
The authors of this study are particularly excited about what their findings could mean for conservation efforts. It is quite challenging to study endangered species for a variety of reasons. Sometimes researchers do not want to disturb an already ailing population; other times, the animals are just elusive and remotely located. However, as climate change and other human-driven stressors affect animal populations across the globe, it is critically important to gain knowledge on endangered populations in threatened habitats. While the technology is still growing and sensors do not yet yield immediate results, the new discovery of using airborne eDNA to identify nearby animal populations will have a vast impact on conservation research.
This new information, although preliminary, exemplifies the rapidly improving advancements to how we are able to gain information from DNA, even at extremely low concentrations in the air. The future might look like a network of monitoring sensors which can continuously collect eDNA to provide information on the location and population size of animals in a remote forest, urban center, or national park. There are several human-related applications of this research as well! In healthcare settings, the air has already been monitored for the DNA of harmful bacteria, viruses, and fungi. However, there are still many unexplored possibilities using airborne eDNA to trace larger organisms. Since DNA can be used to identify individual people, it is not impossible that such a tool could be used in forensics applications or for population-wide disease monitoring in urban settings.
So next time you are on a stroll, remember to stop and smell the roses, but also the kangaroos and zebras and rhinoceroses.
Peer Editors: Yogitha Chareddy & Devan Shell